ECG heartbeat classification¶
This document describes how to classify heartbeats according to its origin.
Description¶
This task implements a heartbeat classifier that follows the EC-57 AAMI recommendation classifying heartbeats into four classes:
- N normal
- S supraventricular
- V ventricular
- F fusion of normal and ventricular
Certain background and introduction to this topic is included in my PhD thesis.
Input Arguments¶
progress_handle — Used to track the progress within your function. [] (default)
progress_handle, is a handle to a progress_bar object, that can be used to track the progress within your function.
tmp_path — The path to store temporary data. tempdir() (default)
Full path to a directory with write privileges.
`payload — A structure to provide audited heartbeat detections to the classifier algorithm. [] (default)
This variable is useful to pass automatic or corrected QRS detections to the classification task. This can be performed as shown in the following example:
cached_filenames = ECGw.GetCahchedFileName({'QRS_corrector' 'QRS_detection'});
ECGw.ECGtaskHandle.payload = load(cached_filenames{1});
mode — Set the classification mode of operation. 'auto' (default)
A control string with any of the following names
- ‘auto’, this mode makes the algorithm operate in automatic mode.
- ‘slightly-assisted’, this mode requires that an expert labels several representative examples, when the algorithm does not reach a confidence level to do it automatically.
- ‘assisted’, this mode is completely assisted. An expert must label all the representative heartbeats from each cluster.
Examples¶
The first example shows the simplest setup of the ECGtask_heartbeat_classifier object, while at the end of this section a complete example with a real signal is shown.
% with the task name
ECG_w.ECGtaskHandle = 'ECG_heartbeat_classifier';
% or create an specific handle to have more control
ECGt = ECGtask_heartbeat_classifier();
and then you are ready to setup the task
% select a mode, automatic mode does not require assistance
ECGt.mode = 'auto';
% this is to use QRS detection previously calculated
cached_filenames = ECG_all_wrappers(ii).GetCahchedFileName({'QRS_corrector' 'QRS_detection'});
ECGt.payload = load(cached_filenames{1})
Finally set the task to the wrapper object, and execute the task.
ECG_w.ECGtaskHandle= ECGt; % set the ECG task
ECG_w.Run();
This example shows in first place, the previous configuration used in recording 208 from MIT Arrhythmia database.
>> ECG_w = ECGwrapper( ...
'recording_name', 'some_path\208', ...
'recording_format', 'MIT', ...
'ECGtaskHandle', 'ECG_heartbeat_classifier', ...
)ECG_w =
############################
# ECGwrapper object config #
############################
+ECG recording: some_path\208 (auto)
+PID: 1/1
+Repetitions: 1
+Partition mode: ECG_overlapped
+Function name: ECG_heartbeat_classifier
+Processed: false
>> ECG_w.Run();
You can follow the evolution in the progress bar, and after a while, it ends and display the classification results
Configuration
-------------
+ Recording: ... \example recordings\208.dat (MIT)
+ Mode: auto (12 clusters, 1 iterations, 75% cluster-presence)
True | Estimated Labels
Labels | Normal Suprav Ventri Unknow| Totals
-----------------|----------------------------|-------
Normal | 1567 6 13 0 | 1586
Supraventricular| 2 0 0 0 | 2
Ventricular | 255 8 1102 0 | 1365
Unknown | 2 0 0 0 | 2
-----------------|----------------------------|-------
Totals | 1826 14 1115 0 | 2955
Balanced Results for
---------------------
| Normal || Supravent || Ventricul || TOTALS |
| Se +P || Se +P || Se +P || Acc | Se | +P |
| 99% 45% || 0% 0% || 81% 99% || 60% | 60% | 48% |
Unbalanced Results for
-----------------------
| Normal || Supravent || Ventricul || TOTALS |
| Se +P || Se +P || Se +P || Acc | Se | +P |
| 99% 86% || 0% 0% || 81% 99% || 90% | 60% | 62% |
This is possible because this recording include the expert annotations, or ‘’ground truth’‘, for each heartbeat. The manual annotations in MIT format are typically included in ‘’.atr’’ files (in this case ‘‘208.atr’‘). Now you can try ‘’slightly-assisted’’ mode, where the algorithm may ask you for help in case of cluster heterogeneity. If this happens, a window like this will appear:
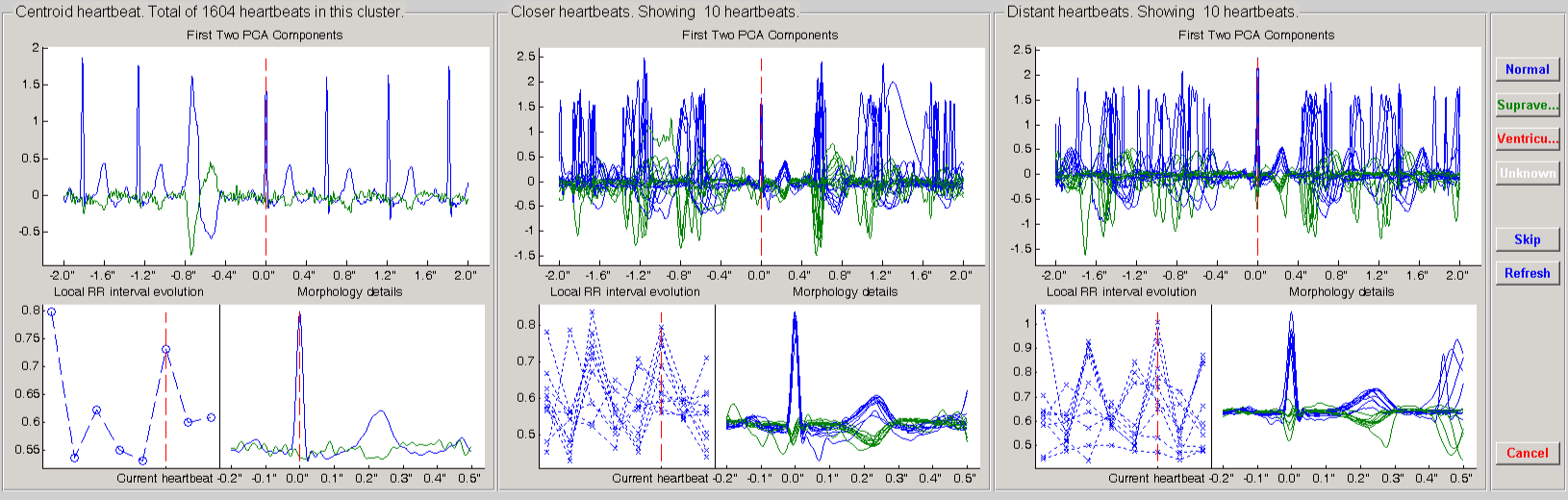
In this window the algorithm is asking you to label the centroid of the cluster, that is showed in the left panel. In the top of each panel some information is showed, as the amount of heartbeats in the current cluster. In the middle panel, you have some examples of heartbeats close to the centroid in a likelihood sense. The same is repeated in the right panel, but with examples far from the centroid. This manner you can have an idea of the dispersion of heartbeats within a cluster. Large differences across the panels indicates large cluster dispersion. If you decide to label the cluster, you can use one of the 4 buttons on your right. The unknown class is reserved for the cases where you can not make a confident decision. At the same time, in the command window, a suggestion appears:
Configuration
-------------
+ Recording: .\example recordings\208.dat (MIT)
+ Mode: assisted (3 clusters, 1 iterations, 75% cluster-presence)
Suggestion: Normal
This means that the centroid heartbeat in the ‘’.atr’’ file is labeled as ‘’Normal’‘. You will see this suggestion for each cluster analyzed, if there are annotations previously available. You are informed about the percentage of heartbeats already labeled with a progress bar, in the bottom of the control panel window.
In case you believe that a cluster includes several classes of heartbeats, you can decide to ‘’skip’’ the classification, and try to re-cluster those heartbeats in the next iteration. You are free to perform as many iterations as you decide, by skipping clusters. The refresh button resamples heartbeats close and far from the centroid, and then redraw the middle and right panels. This feature is useful for large clusters.
You can check the result of this task for every heartbeat in the recording using the visualization functions.
Also check this example for further information.
Results format¶
The results file includes three variables, the annotation type or
classification label anntyp, containing a char label per heartbeat,
which is the initial letter of the heartbeat label. A vector of samples
called time (in correspondence with anntyp), with the occurrence of
each heartbeat labeled in this task. The last variable, is a label list
called lablist, which is a cell array of strings with the full name
of each label in anntyp.
More About¶
Here are some external references about heartbeat classification:
- EC-57 AAMI recommendation
- EP limited software